All along the supply chain, vendors often pass off cheaper fish species as their more valuable counterparts. When one fish fillet looks much like another, misidentification and even deliberate fraud are easy. An EU-funded project has pioneered simple DNA fingerprinting techniques that help consumers get the fish they pay for.
The challenge
When is a hake not a hake? Consumers across Europe, particularly in Spain where the European hake (Merluccius merluccius) is prized, are often deceived into paying higher prices for other species from the hake families (Merluccius and Macruronus). These are all good fish to eat – but often, they are not the European hake consumers think they are buying.
Bluefin tuna is another valuable fish which dishonest traders may substitute with more common varieties. With one fish fillet or steak often looking much like another, mislabelling is easy. It is estimated that nearly 40% of fish sold by fishmongers, supermarkets and restaurants around the world is not what it purports to be.
Confusion and fraud are widespread
At a conference on Greek fisheries science in 2019, seafood processing companies noted that confusion between species is costly for the industry. Shipments often turn out to be the wrong kind of fish – and, perhaps worse, a cargo that is supposed to be made up of one type of fish can be a mixture of several species.
Traders may also need to identify more than just the species. The right type of fish could still come from a different part of the ocean where stocks are less sustainable, or even from a marine reserve area. It might be a farmed fish masquerading as a wild one, or vice versa. DNA testing can help fight fraud by distinguishing between different fish populations, as well as different species.
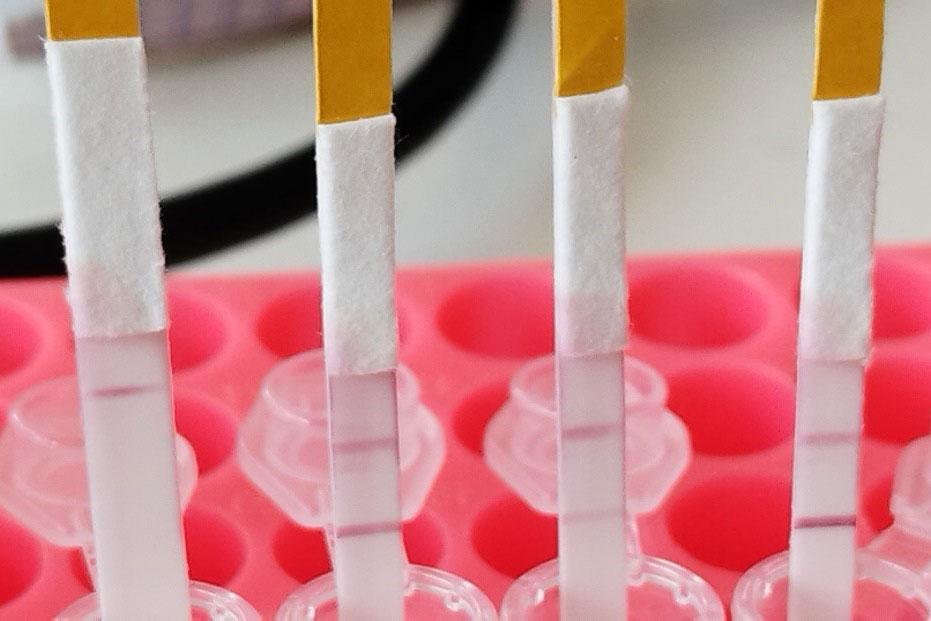
Developing a rapid DNA test
To deal with this problem, scientists agreed to develop a simple DNA test based on a disposable ‘dipstick’, similar to rapid tests for COVID-19. They agreed that the tests should be quick and easy to carry out in the field, and customisable to suit the needs of individual fish processors. Close industry involvement was important to guarantee the success of the project.
The Hellenic Centre for Marine Research, with its extensive experience in the fishing industry, and the University of Patra, which had previously worked on dipstick tests to identify fraudulent products in other sectors such as olive oil, carried out the project.
The scientists developed a set of dipsticks to check the identities of two groups of similar fish species.
- In the first group, they focused on sardines (Sardina pilchardus), which are sometimes confused with European anchovies (Engraulis encrasicolus) and the round sardinella (Sardinella aurita).
- In the second group, it was yellowfin tuna (Thunnus albacares) and skipjack (Katsuwonus pelamis), which are often sold as the more valuable bluefin tuna (Thunnus thynnus).
In addition to the dipsticks, the researchers developed protocols and training materials that allow fish processing companies to positively identify these species in just 90 minutes, without access to a laboratory.
The project received support from the European Maritime and Fisheries Fund (EMFF). EMFF covered all aspects of the work, from laboratory infrastructure to the hands-on training of the industrial partners. The success of the project would not have been possible through other funding mechanisms, the scientists recognised.
Dr Alixis Conides, scientific responsible of the DNASEEKER project, said: “With this project, the beneficiaries had the opportunity to study a very important aspect of DNA application, that is, an accurate and simpler method for species identification with significance for the fish product processing and with more than one application.”
He added: “The applications range from the identification of aquaculture, climate change and alien species; to environmental DNA-based biodiversity studies of marine, brackish and freshwater environments. All of this is done with minimal effort, extremely cheap raw materials, portable instrumentation and a protocol that enables inexperienced scientists or technicians to carry out the analysis with the same results.”
The result is a practical and cost-effective way to rapidly identify fish species that are often mislabelled all along the supply chain, costing consumers money and possibly hindering conservation efforts. Project partners say the technique has the potential for global use and could be expanded to cover other species as necessary.